Habenstein, B., Loquet, A., Giller, K., Vasa, S., Becker, S., Habeck, M., Lange, A.: Hybrid Structure of the Type 1 Pilus of Uropathogenic E. coli. Angewandte Chemie (2015) doi:10.1002/ange.201505065
Shahid, S., Bardiaux, B., Franks, T., Krabben, L., Habeck, M., Rossum, B., Linke, D.: Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nature Methods 9:1212–7 (2012) doi:10.1038/nmeth.2248
Habeck, M.: Bayesian estimation of free energies from equilibrium simulations. Physical Review Letters 109:100601 (2012) doi:10.1103/PhysRevLett.109.100601
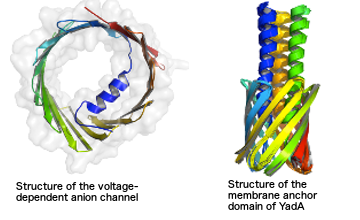
Many cellular processes are governed by macromolecular machines are formed by large complexes of protein and nucleic acids. We develop computational methods to model and characterize these biomolecular structures at various levels of resolution. Using Bayesian statistics we integrate heterogenous experimental data from NMR spectroscopy, cryo-electron microscopy and cross-linking / mass sprectrometry. To apply our approach in practice, we need efficient algorithms to sample all relevant conformational states of the system. Borrowing concepts and methods from statistical physics and machine learning, we have developed and implemented Markov chain Monte Carlo algorithms for probabilistic biomolecular modeling bundled in the Inferential Structure Determination (ISD) software. We have successfully applied ISD in a number of challenging structure determination projects including the structure of the voltage-dependent anion channel from low-resolution X-ray and sparse NMR data, as well as the recent structure of the membrane anchor domain of the trimeric autotransporter adhesin YadA from solid-state NMR data.