Computational Structural Biology
Group Leader
PhD Students
|
• Paul Joubert
|
|
• Ivan Kalev
|
|
• Maren Mahsereci
|
|
• Martin Mechelke
|
Description
Computational structural biology studies aspects of biomolecular structure and dynamics by means of computational methods. The group's focus is on tool development for biomolecular structure determination, prediction and modeling. We are mainly interested in using experimental data to determine biomolecular structures and conformational changes. Our major analytical tool is Bayesian probabilistic inference because it is model-driven, allows the quantification of parameter and model uncertainty, solves inverse problems self-consistently by the use of Bayes's theorem and is capable of integrating data from diverse sources.
Structural information on biological macromolecules can be obtained through a variety of experimental techniques including x-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy and electron microscopy (EM). However with decreasing data quality and quantity, structure determination often becomes a matter of pass or fail. Nonetheless, there is a growing interest in the structural characterization of multi-component molecular machines and membrane proteins which are difficult to crystallize and beyond the size limits of standard NMR experiments. It will therefore be increasingly important to combine diverse experimental data that are themselves not sufficient to fully determine atomic resolution structures and to sample the conformational space that is compatible with all the available structural information. Our goal is to develop methodology and software that can deal consistently with "problematic" structural data including sparse and low-quality data as well as heterogeneous data from diverse experimental sources. We use a Bayesian probabilistic framework to deal with incompleteness and inaccuracy of experimental data and compensate for lack of information by adapting concepts from ab initio structure prediction. We could show that our approach allows us to calculate more accurate structures from limited NMR data and to assess the quality of a structure objectively.

 Structure calculation from sparse experimental data. Left: standard structure calculation, right: Bayesian calculation.
Structure calculation from sparse experimental data. Left: standard structure calculation, right: Bayesian calculation.
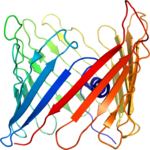
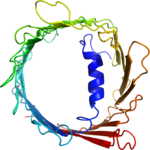 Structure determination of the voltage-dependent anion channel by combining NMR and X-ray crystallographic data.
Structure determination of the voltage-dependent anion channel by combining NMR and X-ray crystallographic data.
Selected Publications
Structure of the human voltage-dependent anion channel.
Bayrhuber M, Meins T,
Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K.
Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15370-5. Epub 2008 Oct 1.
Weighting of experimental evidence in macromolecular structure determination.
Habeck M, Rieping W, Nilges M.
Proc Natl Acad Sci U S A. 2006 Feb 7;103(6):1756-61. Epub 2006 Jan 30.
Inferential structure determination.
Rieping, W.,
Habeck M., Nilges M.
Science 309, 303-6 (07/08/ 2005)



